Sustainability of food side streams: a case study of fermented blends made with sour whey and sunflower press cake powder using the back-slopping technique
- 1Department of Food, Environmental and Nutritional Sciences, Università degli Studi di Milano, Milan, Italy
- 2School of Industrial and Information Engineering, Politecnico di Milano, Milan, Italy
- 3Faculty of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences, Ås, Norway
- 4One Heath Unit, Department of Biomedical, Surgical and Dental Sciences, Università degli Studi di Milano, Milan, Italy
The exploitation of by-products is a key factor to increase the sustainability of the agri-food chain and fermentation is a simple and eco-friendly process for achieving safe and suitable food materials. In this study, we investigated the possibility to manage a spontaneous fermentation of blends made with different proportions of two food side streams (bovine acid whey and sunflower press cake powder) through the application of a back-slopping technique of the mixed material incubated at 26°C in static conditions. A full-factorial 2-factor 3-level design of experiment was applied to infer the effect of the percent (w/w) of press cake powder in the mixture (20, 25, and 30%) and the rate of back-slopping inoculum (15, 30, and 45%). The pH value, titratable acidity, content of sugars, organic acids, and phenolic acids, enumeration of lactic acid bacteria, yeasts and molds, bacterial contaminants, presumptive Bacillus cereus, and Escherichia coli were measured for each fermentation step at 0, 24, 48, and 72 h. On the same samples, a metataxonomics analysis, targeted on bacterial 16S rRNA gene and fungal ITS region, was performed by using the Illumina MiSeq platform. Acidification of the blends (on average, starting pH = 5.45 ± 011, final pH = 4.61 ± 0.11; starting acidity =13.68 ± 1.02 °SH/50 mL, final acidity = 28.17 ± 2.92°SH/50 mL) and high LAB counts (on average, 9.39 log CFU/g ± 0.25) were observed at the end of each refreshment. In all fermented mixtures, B. cereus, E. coli, and molds counts were lower than the detection limit (<2 log CFU/g), whereas bacterial contaminants, overall spore-formers, were always present (3.74 log CFU/g ± 0.27). After 72 h, the dropping of pH value was maximum, yielding significant differences compared to previous fermentation steps (p < 0.01); particularly, the lowest pH (4.45 ± 0.06) was achieved in the central points of DoE (25% of press cake powder and 30% of back-slopping rate), representing the most suitable condition. Results from both culture-dependent and -independent techniques were consistent; although Lactococcus lactis, continuously deriving from the acid whey, was the main LAB, Pediococcus pentosaceus appeared and, in some cases, became the dominant species. Finally, a long-term trial (about 1 month), using the best condition previously pointed out, was performed with an extension of the incubation time to 84 h for each refreshment. The increase in acidity forced the natural selection toward acid-tolerant microbial strains confirming the former results. Although preliminary, these findings can be useful for developing innovative operations to manage these two relevant side streams implementing the circularity of food resources.
1. Introduction
To target the objectives of the ecological transition proclaimed in the act of the European Commission (2019), the enhancement of sustainability in food supply chains is forced to a different use of by-products, especially those containing an attractive level of proteins. This involves rethinking the organization of the food processes, exploiting innovative approaches which should allow connecting material flows that were not previously foreseen and transforming food side streams or wastes into affordable ingredients for new products (Winkler, 2011; Mangieri et al., 2022).
Cheese whey is undoubtedly one of the most interesting by-products due to its content of nutrients and wide spreading of the supply chain (Moates et al., 2016), with an estimated amount of 180 million tons per year (European Commission, 2023). Furthermore, reusing whey is an important challenge for the protection of the environment, being an ecological burden to be disposed of as waste material. Indeed, depending on the enterprise size and the involved business, dairy manufacturers are required to undergo significant economical and logistical efforts for managing it (Risner et al., 2019). Broadly speaking, whey is the liquid derived from the cheesemaking process after curdling, either through enzymatic coagulation or acidification, which causes the precipitation and removal of caseins and fat. In the former case, it is called “sweet” whey, having ~5.0% w/v lactose content, pH > 5.6, variable microbial counts, and almost intact whey proteins (0.9% w/v). This is frequently recycled by large companies as a source for preparing numerous value-added products, such as concentrated whey proteins and lactose, whose production requires huge investments. In the latter case, it is called “acid” or “sour” whey, with pH < 5.0, a lower lactose content (4.2–4.9% w/v), a high cell concentration of lactic acid bacteria (LAB), if fermented by starter cultures, and partially hydrolyzed whey proteins 0.8% (w/v; Ryan and Walsh, 2016). In recent years, acid whey production has increased because of the global growing consumption of Greek-style yogurt and acid-curd cheeses, such as quark cheese, cottage cheese, Petit Suisse, and cream cheese (Luo et al., 2021; Rocha-Mendoza et al., 2021). Due to its less favorable techno-functional properties for further transformation (Risner et al., 2019), the dairy industry and mainly small and medium-sized enterprises (SMEs) still prefer to sell sour whey for piglet feeding or inappropriately discharge it into the sewage system or fields when located in marginal areas (European Commission, 2023). Consequently, from a circular economy perspective, alternative use for this specific dairy side stream is necessary (Rocha-Mendoza et al., 2021).
After Russia, European Union is the second-world area for sunflower oil production with 4.6 million metric tons per year (USDA, 2023). In line with the positive trend toward organic food supply chains, sunflower has the advantage over soybean since its seeds are not genetically modified, making easier its use in organic feed and food chains. In addition, its cultivation can be easily included in crop rotation systems. Obtained after pressing of sunflower seeds for oil extraction, the sunflower press cake is another promising untapped by-product (Moates et al., 2016), being a suitable source of proteins, fibers, unsaturated fats, and polyphenols (González-Pérez and Vereijken, 2007; Anjum et al., 2012). Representing up to 65% of the whole seed dry weight, it generates a considerable amount of waste that is currently used in formulations for animal feeding. Furthermore, the natural presence of anti-nutritional compounds such as phytic and chlorogenic acids can be reduced through fermentation (El Hag et al., 2002), making the sunflower press cake more suitable for human consumption.
Throughout the history of mankind and currently in the less industrially developed areas, fermentation is still the simplest and most convenient operation for enhancing the safety of foods, extending their shelf life, and improving their sensory and nutritional properties (Motarjemi, 2002; Terefe and Augustin, 2020).
In the present study, the optimization for a spontaneous fermentation process of a blend of bovine sour whey, as a liquid component rich in LAB, and powder of ground sunflower press cake as a solid component, is described. For this purpose, a back-slopping technique inspired by traditional sourdough technology (Leroy and De Vuyst, 2004) has been applied: this model practice pointed to drive the transformation of the mixture by exploiting the microbial loads carried by the sour whey to obtain proper acidification of the matter for safety purposes. As in other food preparations such as kefir or sourdough (Özer and Kirmaci, 2014; Von Gastrow et al., 2021), a phenomenon of adaptive evolution can shape and stabilize the microbial community, because of selection pressure exerted by the actual environmental conditions and mutualistic cross-feeding among strains (Londero et al., 2012; Barber et al., 2022). Both culture-dependent and culture-independent approaches have been used to investigate the dynamic of microbial populations grown in the blend samples. In a previous study (Mangieri et al., 2022), Lactococcus lactis strains proved to be the most promising LAB in the fermentation of the mixed material (bovine sweet whey/sunflower press cake). Therefore, we used sour whey from acid-curd cheese manufacturing, in which this species is the dominant one (Foschino et al., 2006). Moreover, from a sustainability perspective, the utilization of acid-curd cheese may allow the reutilization of the LAB already present in the substrate without further addition of bacterial inoculum, saving technical operations and lowering production costs and energy consumption (Rao et al., 2021). The proposed fermented material can be considered newsworthy as a semi-finished product for manufacturers of snack and bakery foods to design innovative and nutritionally valuable products starting from side streams (Raak et al., 2022).
2. Materials and methods
2.1. Design of experiment
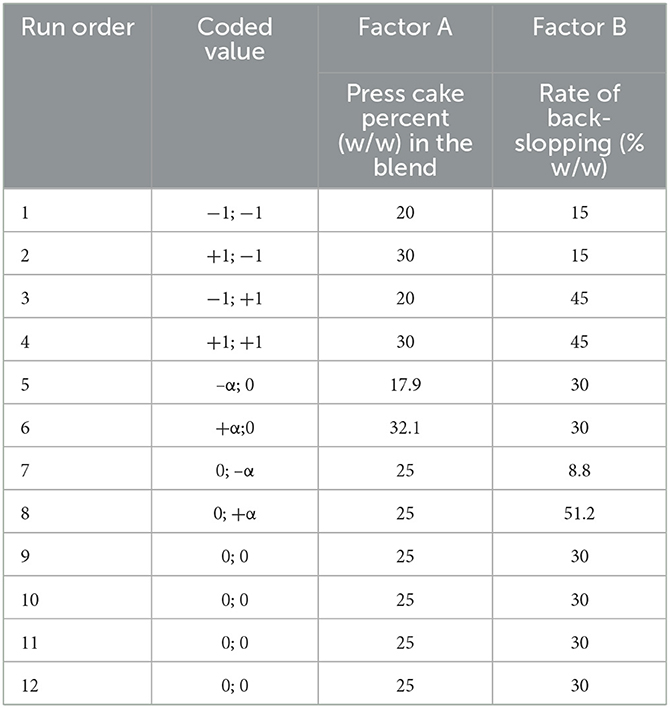
To investigate the effect of different proportions of sunflower press cake powder/sour liquid whey and the rate of back-slopping on the fermentation process, a quadratic model design using the Design-Expert software (v.8.0.7.1, Stat-Ease Inc, Minneapolis, USA) was considered. Each variable, the percentage (w/w) of press cake powder in the blend (Factor A) and the rate of back-slopping inoculum (Factor B), was studied at three different levels (1, 0, +1) adding four axial points, also called “star” points away from the central points by distance ± α, where
The experimental design provided 12 runs, including four tests having the same variable values (Table 1).
Six responses were measured for each fermentation step at 0 (t0h), 24 (t24h), 48 (t48h), and 72 (t72h) h: pH value, enumeration of LAB, fungi (yeasts and molds), bacterial contaminants, presumptive Bacillus cereus, and Escherichia coli. According to the model proposed by the software, the order of the experimental runs was randomized (Table 1). Each test was carried out independently.
2.2. Sample preparation and back-slopping protocol
The powder of organic sunflower press cake, obtained by cold pressing and milling of the whole seeds, was purchased from an external supplier (Ölmühle Moog, Lommatzsch, Germany) and its composition is reported in Supplementary Table 1. The organic acid whey was produced on a lab scale because it was not possible to collect it at local cheese factories in due time for experiment conduction. Moreover, in conventional cheesemaking, potassium sorbate is used as an antifungal to extend the shelf life of fresh acid cheese; this additive might have interfered with the growth of microbial groups. The acid whey was produced by inoculation of a Lactococcus lactis fresh culture (0.5%), previously isolated from Caprino cheese (Emilio Mauri S.p.A., Pasturo, Italy), in 2 L of pasteurized whole milk (3.6% w/v fat content, 3.2% protein, 4.9% lactose) from an organic farm (Centrale del latte di Milano, Milan, Italy) and added with 0.075 g/L of dried calf rennet (Ha-La, Christian Hansen, Hoersholm, Denmark, titer 1:100000). After incubation at 26°C for 15 h, the curd was finely broken and filtered through paper (type Whatman A4). The pH value of the whey was adjusted to 5.0 with NaOH 1M.
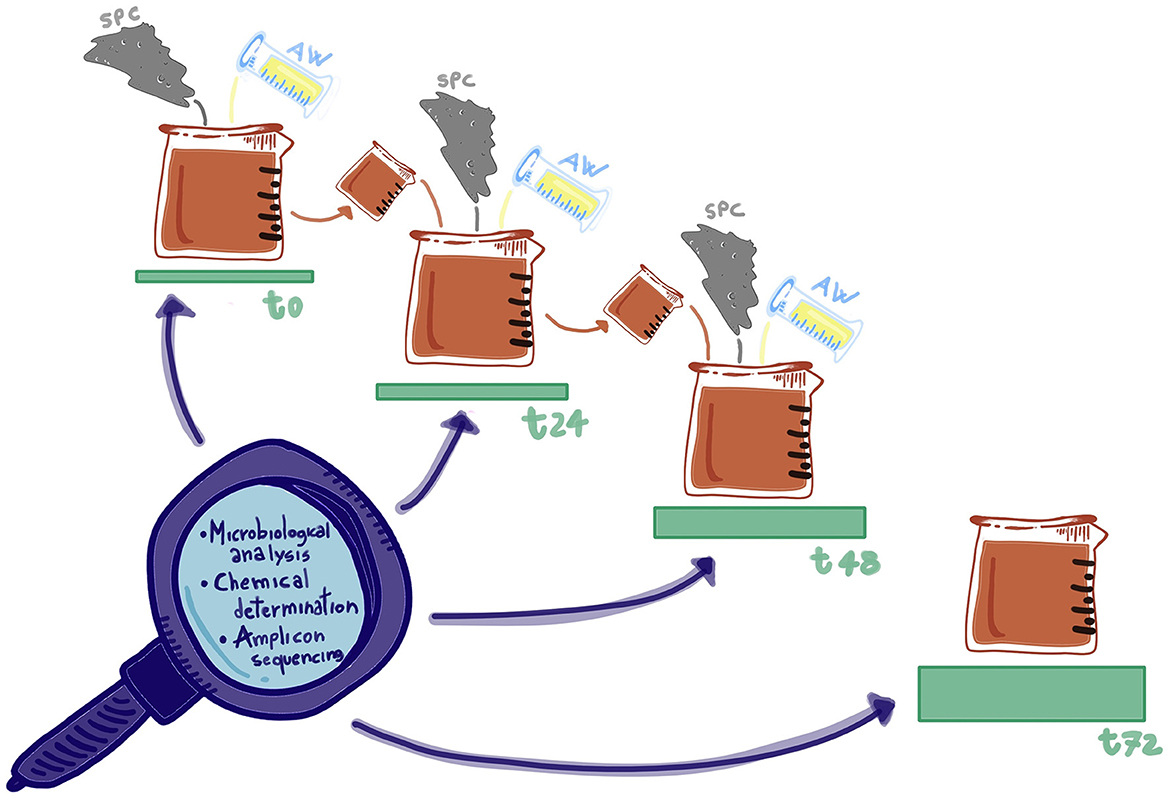
The different blends were prepared as reported in Table 1 (factor A) and homogenized in a mixer (MU, Gerlingen M48CR1/05, Robert Bosch GmbH, Germany) at 95 rpm for 5 min. Then, 200 g of each blend was transferred in a 200-mL sterile jar (Aptaca, Canelli, Italy) and incubated at 26°C in static conditions with a closed screw cap. In this way also, it was taking advantage of the semi-anaerobiosis condition to lessen the growth of aerobic microorganisms by reducing the headspace. After 24 and 48 h, the sample was removed from the container and mixed, as previously described, with fresh press cake powder and acid whey (pH = 5.00) according to the percentage of back-slopping (factor B) indicated in Table 1 and reintroduced again in the jar (Figure 1).
2.3. Microbiological analysis of the blends
At 0 (t0h), 24 (t24h), 48 (t48h), and 72 (t72h) h, samples of the different blends were microbiologically analyzed by plate count technique using the following growth media: Heterofermentative Homofermentative Differential medium (HHD broth, Biolife Italiana srl, Milan, Italy) with added agar (1.8% w/v) for LAB enumeration, plates were incubated at 30°C for 48 h; sugar-free medium (SF agar, Liofilchem srl, Roseto degli Abruzzi, Italy) for aerobic bacterial contaminants and endospores counts, plates were incubated at 30°C for 48 h; in case for endospores count, the samples were previously treated at 80°C for 15 min; Polymyxin B, egg yolk emulsion, Mannitol, and Bromothymol blue medium (PEMBA agar, Scharlab, Sentemenat, Spain) for presumptive enumeration of Bacillus cereus, plates were incubated at 30°C for 48 h; Tryptone Bile X-glucuronide medium (TBX agar, Merck, Darmstadt, Germany) for Escherichia coli counts, plates were incubated at 37°C for 48 h; and Chloramphenicol Glucose Agar (GCA, Scharlab) for yeast and mold counts, plates were incubated at 26°C for 72 h.
2.4. Identification of microbial strains
Some microbial isolates, deriving from colonies randomly collected at the highest dilution in the different media used, were identified. The bacterial DNA extraction was performed through the phenol/chloroform protocol (Green and Sambrook, 2012). Species identification of bacterial isolates was carried out by DNA sequence analysis, targeting on 16S rDNA gene by using universal primers for the amplification of 16S rDNA BSF-8 (5′-AGAGTTTGATCCTGGCTCAG-3′) and BSR-1541 (5′-AAGGAGGTGATCCAGCCGCA-3′; Wilmotte et al., 1993); the relevant amplification conditions were previously reported (Vigentini et al., 2016). The fungal DNA extraction was performed according to Querol et al. (1992). Species identification of fungal isolates was carried out by amplification of the D1/D2 domain 26S rDNA gene by using primers NL1 (5′-GCATATCAATAAGCGGAGGAAAAG- 3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3') as previously described by Kurtzman and Robnett (1998). The PCR products were run in a 1% agarose gel in 0.5% TBE buffer (Tris-borate-EDTA) stained with ethidium bromide (0.4 mg/mL) at 90 V for 1 h and the images were captured with a transilluminator (Biorad, Hercules, California, USA). The PCR amplicons were purified with the EuroClone® spinNAker purification kit (Pero, Milano, Italia) and then sequenced by an external provider (Eurofins Genomics, Vimodrone, Italy). The obtained sequences were compared by Basic Local Alignment Search Tool (BLAST; http://www.ebi.ac.uk/blastall/nucleotide.html) and species identification was attained when homology was more than 98%.
2.5. Chemical analysis of the blends
The different blends were analyzed at times 0, 24, 48, and 72 h. The pH value was measured by a pH meter (Jenway™ 3510 pH meter, Cole-164 Parmer, Stone, United Kingdom).
Acidity was evaluated by potentiometric titration. Samples were titrated with 0.25 N NaOH to pH 8.3 and the titratable acidity was expressed as SH/50 mL. Sugars and organic acids were determined using an HPLC Alliance 2695 pump system (Waters, Milford, MA, USA) equipped with a model 2414 differential refractometer (Waters). The HPLC separation of organic acids was conducted using an Aminex HPX-87H column (300 mm i.d. × 7.8 mm) from Bio-Rad (Segrate, Italy) kept at 50°C. The analytical conditions were as follows: flow 0.6 mL/min, eluent 0.01 N H2SO4, and injection volume 2 μL. Samples were centrifuged at 14,000 g for 20 min and then at 16,100 g for 10 min. Subsequently, 0.5 mL of supernatant was mixed with 2 mL of MilliQ water (Millipore, Darmstadt, Germany) and then filtered through a 0.22-μm pore size membrane filter (Millipore) before injection. The analysis of sugars was carried out using the same HPLC system and detector, but with two Aminex HPX-87P columns in a series (300 mm i.d. × 7.8 mm, Bio-Rad) kept at 75°C. The analytical conditions were as follows: flow 0.6 mL/min, eluent MilliQ water, and injection volume 5 μL. Samples were centrifuged at 14,000 g for 20 min and then at 16,100 g for 10 min. Subsequently, 0.75 mL of supernatant was mixed with 3 mL of Biggs reactive and then brought to volume in a 25-mL flask with MilliQ water. Finally, the samples were filtered through a 0.22-μm pore-size membrane filter (Millipore) before injection. Quantification of organic acids and sugars was performed using aqueous solutions of ethanol and lactic, acetic acids, lactose, sucrose, and raffinose as external standards. All sugars and organic acids were of analytical grade (Sigma-Aldrich, Merck group, Burlington, USA).
The concentrations of caffeic acid, chlorogenic Acid (3-O-caffeoylquinic acid), and its derivatives, namely 4-O-caffeoylquinic acid and di-O-caffeoylquinic acid, were determined by UPLC-UV. An Acquity HClass UPLC (Waters) system equipped with a photodiode array detector 2996 (Waters) was used. The detection wavelength was 320 nm. The separation column was a Kinetex RP18 (150 × 2.1 mm, 2.6 μm, 100 Å; Phenomenex, Torrance, CA, USA) kept at 40°C. The chromatographic separation was carried out using formic acid of 0.2% (v/v) in MilliQ-treated water (solvent A) and methanol (solvent B) as eluting solvents. The UPLC separation was achieved by an elution gradient (2–15% of solvent B in 20 min; from 15 to 50% in 10 min) at a flow rate of 0.6 mL/min. An eight-point calibration curve was obtained for caffeic acid and 3-O-caffeoylquinic acid in the range of 5–3,500 mg/L. The quantification was carried out with the external standard considering caffeic acid and 3-O-caffeoylquinic acid, whose derivatives were quantified as 3-O-caffeoylquinic acid equivalents. Chromatographic data acquisition and processing were performed by Empower 2 software (Waters). The identification of 3-O-caffeoylquinic acid derivatives, namely 4-O-caffeoylquinic acid and di-O-caffeoylquinic acid, was determined by UPLC/ESI-HR-MS working at a spray voltage of −2.5 kV, as described by Palmioli et al. (2019).
2.6. DNA extraction and culture-independent amplicon sequencing
The total DNA from the sample at each time (0, 24, 48, and 72 h) was extracted using the Dneasy PowerSoil Pro kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA samples were stored at −20°C until analysis. 16S rRNA amplicon sequencing was performed with the same procedure described by Skeie et al. (2019). The V3 and V4 regions of the bacterial 16S rRNA were amplified using the method described by Porcellato and Skeie (2016) with minor changes. In brief, 2 μL of DNA was added to the PCR reactions mix containing 1x of Q5 high fidelity master mix (New England Biolabs, Ipswich, MA, USA) and 0.2 mM of each primer. The PCR amplification conditions were similar to those previously reported (Porcellato and Skeie, 2016). The ITS region of the fungal operon was amplified according to the method described by Østlie et al. (2021). After PCR amplification, the PCR product was purified using 0.6X of Agencourt AMPure XP beads (Beckman Coulter, Inc, Brea, CA, USA), according to the manufacturer's instructions. In total, 5 μL of the purified PCR product was used as a template for the second PCR using primers with unique 8 bp barcodes. The PCR reaction was similar to the previous PCR and the amplification was performed as follows: initial denaturation at 98°C for 30 s and 10 cycles of denaturation for 15 s, annealing for 30 s at 55°C, and elongation for 20 s at 72°C. Final elongation was performed for 5 min at 72°C. The PCR product was then purified and normalized using the SequalPrep Normalization Plate Kit (Thermo Fisher Scientific, Waltham, MA, USA). The purified library was quantified using the KAPA library quantification kit (KAPA Biosystems, Wilmington, MA, USA) and sequenced on an Illumina Miseq platform (Illumina Inc., San Diego, CA, USA) using the Miseq Reagent kit v3 (Illumina Inc.). During library prep for the 16S and ITS, the same indices were used for both amplicons and the available FASTQ files contains sequence from both of them. The raw reads have been deposited at the European Nucleotide Archive with accession number PRJEB54064.
2.7. Bioinformatic analysis from amplicon sequencing
During bioinformatics, analysis reads were quality filtered and trimmed using the Dada2 package using truncating of forward reads set to 265 bases and truncating of reverse reads set to 220 bases (Callahan et al., 2016). The error model in Dada2 was created using 1 million random filtered reads. Sequence variants (SV) were inferred using the DADA2 algorithm and the removal of chimeras was performed using the function “removeBimeraDenovo” in the Dada2 R package (Callahan et al., 2016). Sequence variants shorter than 375 and 250 base pairs for bacterial 16S and ITS, respectively, were removed from the final table. Taxonomy was assigned using the Decipher R package against the SILVA SSU database and the UNITE database for bacterial 16S and ITS sequence, respectively (Quast et al., 2013; Wright, 2015). An alpha and beta diversity analysis was performed using the R package vegan (Oksanen et al., 2017). Bray–Curtis dissimilarity matrixes were selected as input for ordination analysis using the principal coordinate analysis using the R package APE and the function EnvFit from the “Vegan” package to fit all the SVs to the ordination plot.
2.8. Statistical analysis
Regarding the results from cultivation techniques, the ANOVA of the data was elaborated with Statgraphics Centurion (v. 18, Statistical Graphics Corp., Herndon, VA, United States); Tukey's HSD test was used to compare the sample means to evaluate significant differences among mean values of main factors. The Pearson correlation was calculated for the caffeic acid, 3-O-caffeoylquinic acid, 4-O-caffeoylquinic acid, and di-O-caffeoylquinic acid considering the critical values (df = 45) of 0.243 (α = 0.1), 0.288 (α = 0.05), and 0.372 (α = 0.01).
3. Results
3.1. Effect of sunflower press cake powder percentage and back-slopping rate on blend fermentation
The microbiological counts (expressed as mean log CFU/g ± standard deviation) of the press cake powder samples before blending and fermentation trials were the following: bacterial contaminants, 4.81 ± 0.36, of which endospores 4.30 ± 0.63; molds, 2.94 ± 0.15; yeasts, < 2; presumptive B. cereus < 2; E. coli, < 2. These data were compared with those found by Podravac et al. (2018) in a survey on the microbiological quality of oilseed products from organic production. Furthermore, the fresh sour whey samples showed an average LAB count of 9.00 log CFU/g ± 0.44, whereas the other investigated microbial groups' results were always below the limit of detection. The pH value was 4.71 ± 0.37 on average. Similar results were obtained by Demarigny et al. (2011) who were studying the influence of the back-slopping practice on lactococci diversity in model cheesemaking.
The data of pH measurements and plate counts obtained at starting and final points after fermentation for each refreshment were inserted into the Design-Expert software. It gave back effect (factors) analysis, ANOVA, and estimated response surfaces. All prepared specimens supported microbial growth, and the acidification of the blends was always observed (Supplementary Tables 2, 3). Regarding the occurrence of potential pathogen bacteria, no sample showed any presumptive colony of B. cereus or E. coli in the corresponding medium plates (< 2 log CFU/g). Likewise, no mold colony was detected in the relevant selective agar medium for any trial (< 2 log CFU/g). The presence of yeasts was noted three times at t24h (runs 2, 6, and 8) and four times at t72h (runs 1, 3, 5, and 7), while in all the other cases, it was under the limit of detection (Supplementary Table 2). The few and unevenly distributed numbers of yeast colonies did not allow us to determine if there were significant differences among the samples. The enumeration of LAB exhibited high values for all experimental runs: at the beginning and after 24, 48, and 72 h, the average count remained stable with values of 9.00 log CFU/g ± 0.36, 9.47 ± 0.25, 9.33 ± 0.24, and 9.37 ± 0.25, respectively. LAB cell concentrations were evaluated through the Design-Expert software, which returned that there were not any significant differences among the different samples (data not shown). It should be considered that, according to the back-slopping rate previously set, for each refreshment, a quantity of fresh sour whey containing a high cell concentration of the used starter culture (on average, 9.00 log CFU/g ± 0.44) was added each time. Bacterial contaminants throughout the fermentation of all samples showed an average value constantly under 4 log CFU/g, with 3.90 log CFU/g ± 0.38, 3.90 ± 0.28, 3.79 ± 0.15, and 3.74 ± 0.23 at t0h, t24h, t48h, and t72h, respectively. Regarding these results, the shape of the response surface obtained for press cake powder percentage and back-slopping rate interaction (Figure 2) at the end of the first refreshment (t24h) revealed that the larger the percentage of press cake in the blends and the mass of re-inoculation for the refreshment, the more bacterial counts grew. It is probably due to a higher microbial load carried back by the plant material and its greater buffering power. When the sunflower press cake powder portion in the mixtures was lower, the counts of bacterial contaminants were more limited and the amount of material used in the back-slopping became not relevant. With the progress of the refreshment steps, the effect was lost and the model did not return a curved response surface after 48 and 72 h of fermentation.
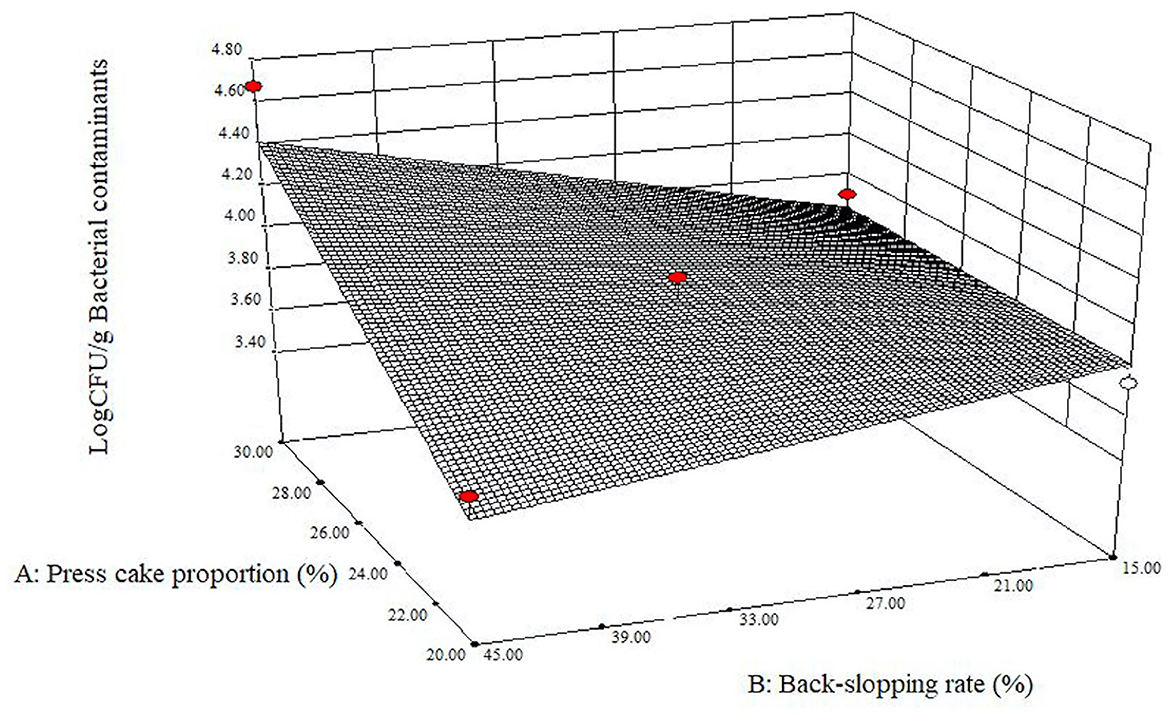
Figure 2. Estimated response surface fitted to experimental data points corresponding to log CFU/g bacterial contaminants at 24 h as a function of press cake percentage and back-slopping rate.
While developing a new material, an important parameter to monitor as a reference point for food safety purposes is the rate of acidification and the attained final pH value. Before fermentation, the pH values of the different blends ranged from 5.25, for the sample with the lowest amount of added press cake powder (run 5), to 5.62, for the sample with the highest percentage of press cake powder (run 6), with an overall average of 5.45 ± 011. After fermentation, the pH values of all samples presented an overall average of 4.61 ± 0.11, with mean values of 4.64 ± 0.09, 4.65 ± 0.10, and 4.55 ± 0.13 at t24h, t48h, and t72h, respectively. The titratable acidity of the different mixtures started from 13.68 ± 1.02 °SH/50 mL to achieve values of 26.72 ± 1.02, 27.64 ± 1.79, and 30.16 ± 3.50 °SH/50 mL at t24h, t48h, and t72h, respectively (Supplementary Table 3). The amounts of lactic acid, which represented the main found organic acid, at the end of each refreshment step were 1.59 ± 0.22, 1.62 ± 0.22, and 1.78 ± 0.46 (g/100 mL) at t24h, t48h, and t72h, respectively. Regarding the achieved pH values, after the third refreshment (t72h), the model is fitted with the central points which caused convexity of the response surface (Figure 3). The curvature arose from the significance of the two quadratic terms (p < 0.01) in correspondence to 25% of press cake proportion (factor A) and 30% of back-slopping rate (factor B), with the lowest pH mean value of 4.45 ± 0.06. It can be highlighted that the highest pH value (4.81) was found at the high level of factor A (run 6, 32.1%), while the lowest one (4.37) was in the central point (run 12).
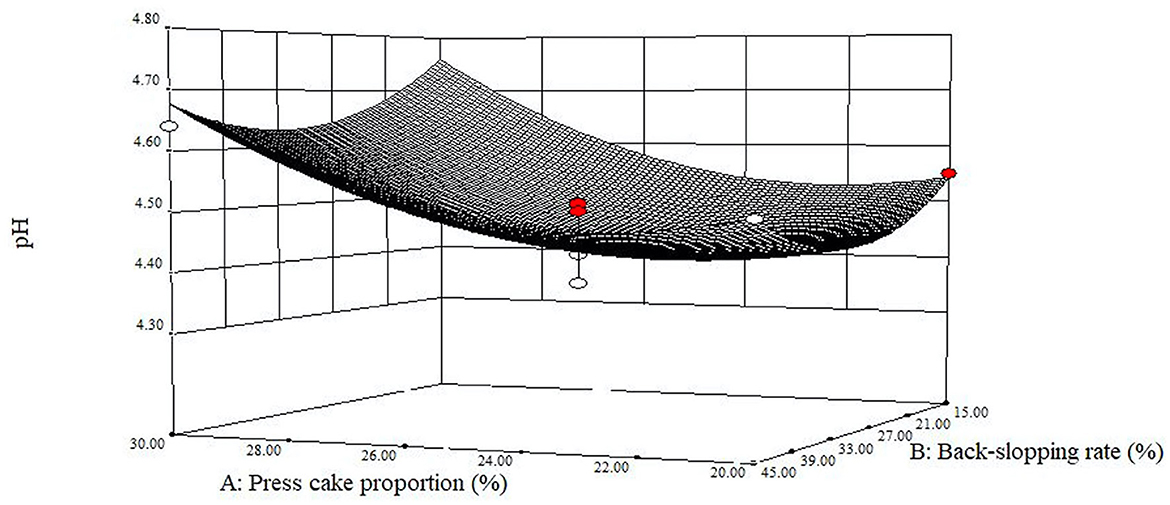
Figure 3. Estimated response surface fitted to experimental data points corresponding to the final pH of the fermented blend at 72 h as a function of press cake percentage and back-slopping rate.
With regard to the evolution of caffeic acid, chlorogenic acid, and its derivatives, the major change occurred within 24 h of fermentation (Supplementary Table 3). Up to 72 h of fermentation, only slight or negligible variations of the investigated phenolic acids were found. While the trend of 3-O-caffeoylquinic was variable depending on the different conditions of fermentation, the content of caffeic acid increased except for runs 3 and 5. The concentration of the two 3-O-caffeoylquinic acid derivatives, 4-O-caffeoylquinic acid and di-O-caffeoylquinic acid decreased during the fermentation, except for runs 1 and 2, showing a significant positive correlation (0.935, α = 0.01). Nonetheless, their decrease (in moles) cannot be explained by the increase of caffeic acid (in mole) even if a negative significant correlation was evidenced with di-O-caffeoylquinic acid (−0.272, α = 0.1). Moreover, a significant positive correlation was found between 3-O-caffeoylquinic acid and 4-O-caffeoylquinic acid (0.322, α = 0.05).
3.2. Species identification of isolated strains
Microbial colonies grown at the highest concentrations in the plates of different culture media were randomly collected after the first (t24h) and the third (t72h) rounds, purified in the same medium by a double consecutive streaking, and subjected to identification by sequencing of the specific target after DNA extraction.
As expected, most of the LAB isolates were ascribed to L. lactis. However, in run 4 at t72h Pediococcus pentosaceus became the dominant species. Regarding the bacterial contaminants, Acinetobacter radioresistens, Bacillus safensis, and Staphylococcus xylosus were the species isolated at t24h in runs 9, 2, and 4, respectively. Among the yeast isolates at t72h, Rhodotorula mucilaginosa was found as the dominant species in run 1 while Saccharomyces cerevisiae was in run 5.
3.3. Metataxonomics analysis
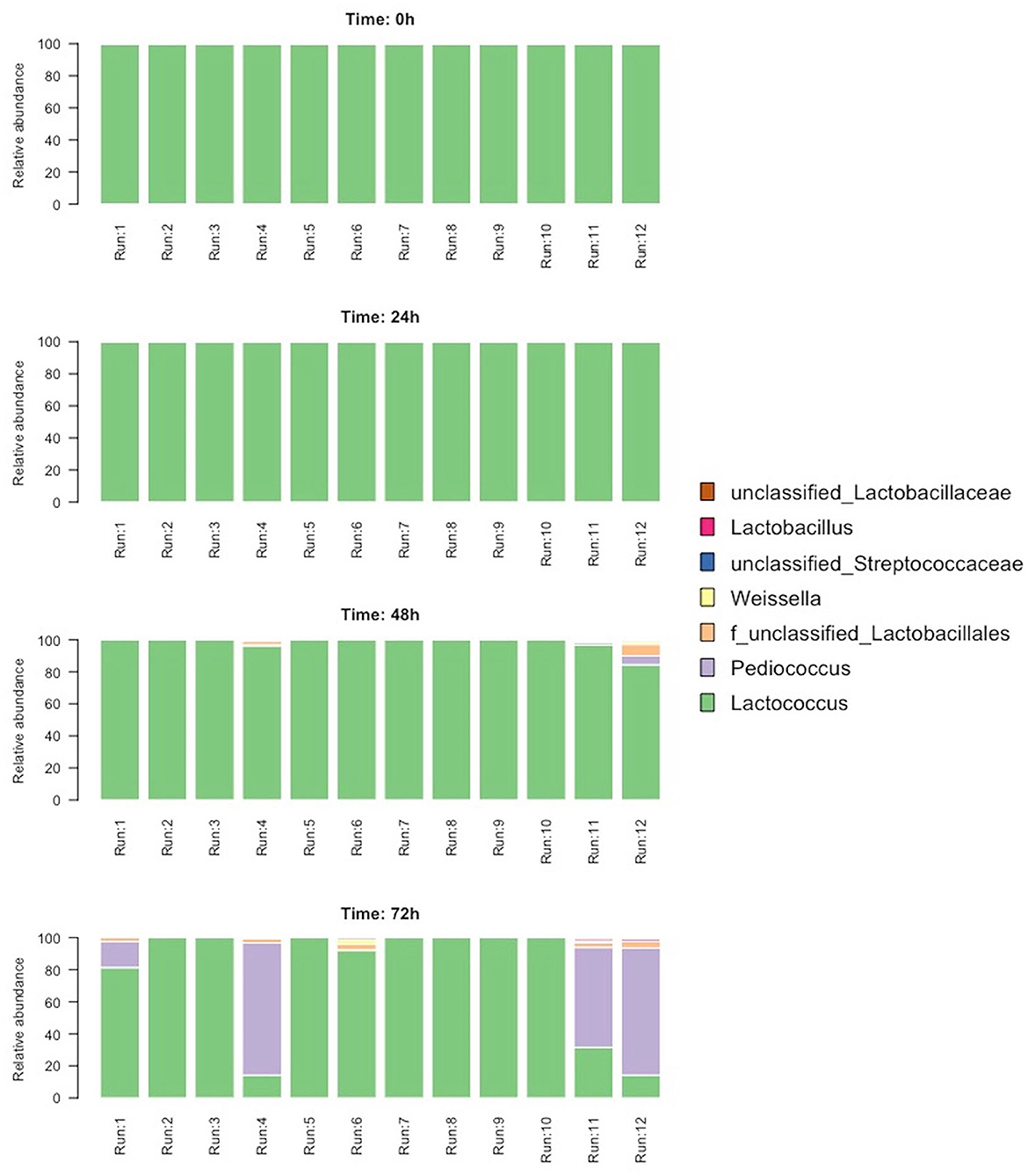
To evaluate the temporal changes in the microbial composition of the blends, amplicon sequencing of all the samples was performed. A total of 544,315 high-quality bacterial reads were obtained from 48 samples (average 11,108 and median 10,305 per sample). The microbiota was dominated by one sequence variant (SV1) belonging to Lactococcus (94.6% of the total sequences). At time 0 and time 24 h, all the samples contained 100% of Lactococcus, whereas, after 48 and 72 h, other SVs were detected (Figure 4). These were taxonomically assigned to Pediococcus, Lactococcus, and unclassified Lactobacillales. Significant changes in microbiota were detected after 72 h, where Pediococcus became the dominant genus (>50% in relative abundance) in samples from runs 4, 11, and 12 (Figure 4). In addition, runs 11 and 12 had a higher microbial diversity (both Chao1 and Shannon indices) compared to the other runs; this increase was already detected after 48 h (data not shown).
After amplification and sequencing of the ITS spacer, a high number of sequences were obtained but after taxonomical assignment against the UNITE database and filtering of eukaryotic sequences, few sequences were obtained per sample (average 136 sequences/sample, median 61). These sequences were assigned to the family Pleosporaceae, Leptosphaeriaceae, Rhizopodaceae, and Saccharomycetaceae. While the first three families contain species of fungus known to be plant pathogens, one SV was detected as Saccharomycetaceae and assigned to the genus Saccharomyces. This SV was present in run 5 at 72 h and confirmed the results obtained with the culture-dependent technique.
3.4. Assessment of a long-term fermentation
To implement the best conditions for acidification obtained from the outcome of the DoE (percentage of press cake powder in the blend = 25 %; rate of back-slopping = 30%), a proof of concept was carried out in a long-term fermentation trial. To avoid the development of harmful bacteria, the acidification of the blend was forced by lengthening the fermentation time. In preliminary results (data not shown), the mixture attained a pH value of < 4.50 in 84 h, on average; subsequently, this time frame was assumed for each step of the back-slopping technique as reported in paragraph 2.2. After homogenization at 95 rpm for 5 min, 200 g of the mixture was transferred in a 200-mL sterile jar and incubated at 26°C in static conditions with a closed screw cap.
As shown earlier, in the first three refreshments, fresh sour whey was used for preparing the blends. From the fourth back-slopping step onward, this was substituted by a sweet whey powder (Flowhey, Lactalis ingredients, Bourgbarre, France), reconstituted at 6% (w/v) and heat treated according to Mangieri et al. (2022) to halt the continuous addition of LAB cells to the material. Samples were collected every 84 h for 28 days (t0h, t84h, t168h, t252h, t336h, t420h, t504h, t588h, and t672h) and microbiological and chemical analyses were performed as described in paragraph 2.5. The colonies found at the higher dilutions were isolated and identified as above mentioned (paragraph 2.4).
This experiment allowed examining the evolution of the microbial population and the stability of the substrate by applying the back-slopping technique by taking on the pH as a limiting factor for the selective pressure on microorganisms. Noteworthy, along the time span of the trial, fungal and bacterial contaminants remained under 2.5 and 4 log10 CFU/g, respectively. The pH of the blend started from 5.33 and after two back-slopping steps (t252h), its value stabilized at 3.94 ± 0.02 until the end of the experiment (Table 2), while the value of titratable acidity raised to 44.9 SH/50 mL ± 5.8. During the 28 days, the LAB always showed high levels of cell concentration between 9.04 and 9.60 log10CFU/g, exhibiting the capacity to grow well and dominate the fermentation process even after t336h when the sour whey was replaced by the sweet whey. At each back-slopping step, colonies of LAB at higher dilutions were randomly isolated from HHD agar plates and identified as previously reported. Interestingly, from time t0h to time t168h, one single type of colony was found that was subsequently ascribed to L. lactis, as expected, being the predominant species in sour whey. From time t336h to the end of the trial, another type of colony became the main morphology onto HHD agar plates and this was identified as P. pentosaceus. A strain of this species was already recognized as dominant in some samples of the DoE test.

Table 2. Results of microbiological and chemical analyses for each step of back-slopping in the long-term fermentation trial.
4. Discussion
The European Commission aims to be carbon-neutral by 2050, moving the actual economy toward a more sustainable system (European Commission, 2019). Undoubtedly, this goal passes from the reuse of food side streams especially those rich in nutrients, such as sour whey and sunflower press cake. Due to the limitation of production during harvest time, sunflowers must be stabilized even when used for animal feeding. For centuries, fermentation is applied for extending the shelf life of raw foods (Motarjemi, 2002). In particular, during a fermentative process where LAB become the dominant population, enzymes and metabolites, such as organic acids, aldehydes, bacteriocins, and antifungal peptides are produced by the bacterial cells, and they transform and stabilize the substrate by preventing or slowing down the proliferation of pathogen and spoilage microorganisms (Siedler et al., 2019; Terefe and Augustin, 2020). In this study, a spontaneous fermentation of side streams blend was investigated through a full-factorial 2-factor 3-level DoE for examining the effect of different proportions of solid/liquid parts and different ratios of back-slopping. The materials used were sunflower press cake powder, carrying a “natural” unavoidable contamination of bacteria and molds, and fresh sour whey deriving from acid-curd cheesemaking, at a laboratory scale, with a high concentration of lactococci, which have been thought as an affordable and suitable microbial inoculum for the blend. In a previous study by Mangieri et al. (2022), strains belonging to L. lactis proved to be reliable for the fermentation of mixtures of similar by-products. Although it was not possible to highlight significant differences among the tested conditions in the different runs of the DoE, LAB was always able to acidify the blends and restrain or decrease the counts of bacterial contaminants as well as those of the molds. Main bacterial contamination can be ascribed to Bacillaceae which form endospores capable to survive for a long period and can be induced into germination by environmental stresses such as nutrient deprivation and changes in water content and temperature. Previous studies detected the presence of spore-forming bacteria (such as Bacillaceae) on the microbial composition of similar matrices (Mandic-Mulec et al., 2015; Jmeii et al., 2019). Considering the lowering of pH observed in all samples, the acidification of the medium played a role to prevent the growth of the possible vegetative forms, while the endospores remained stable. As well-known, the pH of a food product is crucial to ensure its safety, and the data analysis of this parameter revealed that the applied model found significant differences among the investigated formulations and conditions, even though at least three refreshments were needed to reduce the pH below 4.5. In particular, this goal was achieved at the end of the fermentation of the third back-slopping step (t72h) in the central positions of the generated response surface for both factors (30% of press cake and 25% as rate of back-slopping).
The relevant content of chlorogenic acid in sunflower press cake was already reported to represent about 1–4% by mass (Scharlack et al., 2017). A clear impact of the microbiota on the evolution of the phenolic compounds determined in this study was not evidenced, suggesting that the bacterial strains present were not able to significantly decrease the 3-O-caffeoylquinic acid and its derivatives in the adopted experimental conditions. The esterase activity of lactic acid bacteria was found in other studies for Lb. gasseri and B. animalis subsp. lactis that showed degradation of chlorogenic acid (Couteau et al., 2001; Raimondi et al., 2015; Fritsch et al., 2016). Other bacteria, such as Lb. plantarum, did show only limited consumption of 3-O-caffeoylquinic acid, while no decrease was observed for P. pentosaceus (Fritsch et al., 2016), in accordance with our results.
The evolution of the microbiota into the blends was also examined through an amplicon sequencing approach. As expected, Lactococcus dominated the microbiota as it was the main bacterium of the starter culture deriving from the sour whey. Surprisingly, at the end of the third fermentation step (t72h) in runs 4, 11, and 12, which revealed the lowest pH values, the genus Pediococcus overcame Lactococcus, despite the latter being continuously supplied through the refreshment steps. The identification of LAB strains performed by the 16S RNA gene sequencing confirmed the results of the culture-independent analysis, since L. lactis proved to be the most frequently isolated species, whereas a P. pentosaceus strain appeared predominant in the run 4.
The experiment of long-term fermentation gave evidence that the decrease of pH value in the substrate operated an effective selection on microorganisms, both hampering the growth of spoilage bacteria and molds and driving LAB strains to become dominant. Particularly, lactococci were the main population until they were continuously added from the portion of sour whey with the back-slopping operation, whereas pediococci took over when the whey type changed. Then, if the environmental and working conditions are permanent as occurs when a back-slopping practice is adopted, the fermentation process stabilizes, without affecting the achievement of the safety level for the obtained mixture, and paves the way for the development of new food material. Further studies are needed to investigate how P. pentosaceus can overcome L. lactis, while it is meaningful to have observed the same phenomenon in similar circumstances, that allows unveiling that the former bacterium exhibited a probably higher growth rate or a greater resistance to the acidity. P. pentosaceus, presumably part of the wild microbiota present in the sunflower press cake powder samples, is frequently found in vegetable fermented foods (Qi et al., 2021) and the interest in it is growing because of its potential probiotic properties (Jiang et al., 2021).
To sustain the hypothesis that pediococci could originate from sunflower, a supplementary analysis of the material enriched and fermented without whey was performed; this trial highlighted the presence of strains ascribed to P. pentosaceus and P. acidilactici (data not shown), agreeing with the previous observation.
The outcome of this study highlights the possibility of managing the microbial acidification of blends of sour whey and sunflower press cake powder by tuning the proportion of the investigated by-products, without compromising safety and effectively counteracting the growth of unwanted microorganisms. Nevertheless, the required process time might be too long for industrial applications, even if the fermentation would improve the material from a nutritional and technological point of view in anticipation of subsequent treatments (Tangyu et al., 2019; Petraru et al., 2021). Moving toward no longer dispensable circular economy system, side streams with a high protein content should be valorized by simplifying the processes (Oliveira Filho and Egea, 2021), especially for SMEs to transform by-products into innovative and sustainable food materials for providing additional remuneration. The transition to a carbon-neutral food industry requires a proper risks/benefits assessment and should be facilitated by updated legislation that meets the needs of the sector (Rao et al., 2021). In the current worldwide scenario of food production, the co-processing of mixed food waste streams is by the application of a simple and cheap technique, as shown in this study through a back-slopping fermentation protocol (Terefe and Augustin, 2020). Our study underlined the potential of this application in limiting the development of microbial contaminants being a reliable solution to extend the shelf life of the side streams. Finally, this material may be considered as a platform for the production of novel food products though the suitability of this process on a larger scale should be further investigated in terms of functional and sensory aspects.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJEB54064.
Author contributions
NM and RF: conceptualization. DP, DF, ID, and IV: methodology. NM, GR, AW, and DF: investigation. NM, GR, DP, AW, and DF: formal analysis. NM, GR, DP, and RF: writing—original draft preparation. NM, DP, ID, DF, RF, and IV: writing—review and editing. RF: supervision, project administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge the financial support for this project provided by Transnational Funding Bodies [the Italian Ministry of Agricultural, Food and Forestry and Tourism Policies (MIPAAF)] and H2020 ERA-NETs SUSFOOD2 and CORE Organic Cofund, under the Joint SUSFOOD2/CORE Organic Call 2019 Project ID 25 FERBLEND: https://susfood-db-era.net/main/content/Joint-call-2019.
Acknowledgments
The authors would like to thank all the partners of the Consortium for supporting our study, particularly the German group coordinated by Harald Rohm for providing materials and relevant composition, and Miss Federica Campo for drawing Figure 1. The authors also acknowledge support from the University of Milan through the APC initiative.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1166002/full#supplementary-material
References
Anjum, F. M., Nadeem, M., Issa Khan, M., and Hussain, S. (2012). Nutritional and therapeutic potential of sunflower seeds: A review. Brit. Food J. 114, 544–552. doi: 10.1108/00070701211219559
Barber, J. N., Nicholson, L. C., Woods, L. C., Judd, L. M., Sezmis, A. L., Hawkey, A., et al. (2022). Species interactions constrain adaptation and preserve ecological stability in an experimental microbial community. ISME J. 16, 1442–1452. doi: 10.1038/s41396-022-01191-1
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Couteau, D., McCartney, A. L., Gibson, G. R., Williamson, G., and Faulds, C. B. (2001). Isolation and characterization of human colonic bacteria able to hydrolyse chlorogenic acid. J. Appl. Microbiol. 90, 873–881. doi: 10.1046/j.1365-2672.2001.01316.x
Demarigny, Y., Dalmasso, M., Tonleu, A., Rigobello, V., Beuvier, E., Ly-Chatain, M. H., et al. (2011). Influence of the backslopping practice on the microbial diversity of the Lactococcus population in a model cheesemaking. Food Nutr. Sci. 2, 618–627. doi: 10.4236/fns.2011.26087
El Hag, M. E., El Tinay, A. H., and Yousif, N. E. (2002). Effect of fermentation and dehulling on starch, total polyphenols, phytic acid content and in vitro protein digestibility of pearl millet. Food Chem. 77, 193–196. doi: 10.1016/S0308-8146(01)00336-3
European Commission (2019). Communication From the Commission to the European Parliament, the European Council, the Council, the European Economic and Social Committee and the Committee of the Regions. The European Green Deal. Bruxelles 11.12.2019 COM (2019) 640 Final.
European Commission (2023). Life for Acid Whey. Reuse of Waste Acid Whey for Extraction of High Added Value Bioactive Proteins. Reference: LIFE16 ENV/SI/000335 Website Visited on 20th March 2023. European Commission LIFE Programme. Available online at: http://lifeforacidwhey.arhel.si/en (accessed February 14, 2023).
Foschino, R., Picozzi, C., Borghi, M., Cerliani, M. C., and Cresci, E. (2006). Investigation on the microflora of Caprino lombardo cheese from raw goat milk. Ital. J. Food Sci. 18, 33–49.
Fritsch, C., Heinrich, V., Vogel, R. F., and Toelstede, S. (2016). Phenolic acid degradation potential and growth behaviour of lactic acid bacteria in sunflower substrates. Food Microbiol. 57, 178–186. doi: 10.1016/j.fm.2016.03.003
González-Pérez, S., and Vereijken, J. M. (2007). Sunflower proteins: Overview of their physicochemical, structural and functional properties. J. Sci. Food Agric. 87, 2173–2191. doi: 10.1002/jsfa.2971
Green, M. R., and Sambrook, J. (2012). Molecular Cloning: Laboratory Manual, 4th edn. New York, NY: Cold Spring Harbor Laboratory Press.
Jiang, S., Cai, L., Lv, L., and Li, L. (2021). Pediococcus pentosaceus, a future additive or probiotic candidate. Microb. Cell. Fact. 20, 45. doi: 10.1186/s12934-021-01537-y
Jmeii, L., Soufi, L., Abid, N., Mahjoubi, M., Roussos, S., Ouzari, H. I., et al. (2019). Assessment of biotechnological potentials of strains isolated from repasso olive pomace in Tunisia. Ann. Microbiol. 69, 1177–1190. doi: 10.1007/s13213-019-01499-y
Kurtzman, C. P., and Robnett, C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73, 331–371. doi: 10.1023/A:1001761008817
Leroy, F., and De Vuyst, L. (2004). Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 15, 67–78. doi: 10.1016/j.tifs.2003.09.004
Londero, A., Hamet, M. F., De Antoni, G. L., Garrote, G. L., and Abraham, A. G. (2012). Kefir grains as a starter for whey fermentation at different temperatures: chemical and microbiological characterisation. J. Dairy Res. 79, 262–271. doi: 10.1017/S0022029912000179
Luo, S., DeMarsh, T. A., deRiancho, D., Stelick, A., and Alcaine, S. D. (2021). Characterization of the fermentation and sensory profiles of novel yeast-fermented acid whey beverages. Foods 10, 1204. doi: 10.3390/foods10061204
Mandic-Mulec, I., Stefanic, P., and Van Elsas, J. D. (2015). Ecology of bacillaceae. Microbiol. Spectrum. 3, TBS-0017-2013. doi: 10.1128/microbiolspec.TBS-0017-2013
Mangieri, N., Ambrosini, D., Baroffio, S., Vigentini, I., Foschino, R., and De Noni, I. (2022). Valorisation of bovine sweet whey and sunflower press cake blend through controlled fermentation as platform for innovative food materials. Foods 11, 1417. doi: 10.3390/foods11101417
Moates, G., Sweet, N., Bygrave, K., and Waldron, K. (2016). Top 20 Food Waste Streams. REFRESH Project Deliverable, 6.9 WRAP/IFR. Available online at: https://eu-refresh.org/top-20-food-waste-streams.html (accessed February 14, 2023).
Motarjemi, Y. (2002). Impact of small-scale fermentation technology on food safety in developing countries. Int. J. Food Microbiol. 75, 213–229. doi: 10.1016/S0168-1605(01)00709-7
Oksanen, F. J., Simpson, G. L., Guillaume Blanchet, F., Kindt, R., Legendre, P., et al (2017). Vegan: Community Ecology Package. R package Version 2.4-3. Available online at: https://CRAN.R-Project.org/package=vegan (accessed February 14, 2023).
Oliveira Filho, J. G., and Egea, M. B. (2021). Sunflower seed by-product and its fractions for food application: An attempt to improve the sustainability of the oil process. J. Food Sci. 86, 1497–1510. doi: 10.1111/1750-3841.15719
Østlie, H. M., Porcellato, D., Kvam, G., and Wicklund, T. (2021). Investigation of the microbiota associated with ungerminated and germinated Norwegian barley cultivars with focus on lactic acid bacteria. Int. J. Food Microbiol. 341, 109059. doi: 10.1016/j.ijfoodmicro.2021.109059
Özer, B., and Kirmaci, H. A. (2014). “Fermented milks, products of eastern Europe and Asia,” in Encyclopedia of Food Microbiology, 2nd edn, eds C. A. Batt and M. L. Tortorello (Cambridge, MA: Academic Press), 900–907. doi: 10.1016/B978-0-12-384730-0.00123-3
Palmioli, A., Bertuzzi, S., De Luigi, A., Colombo, L., La Ferla, B., Salmona, M., et al. (2019). BioNMR-based identification of natural anti-Aβ compounds in Peucedanum ostruthium. Bioorg. Chem. 83, 76–86. doi: 10.1016/j.bioorg.2018.10.016
Petraru, A., Ursachi, F., and Amariei, S. (2021). Nutritional characteristics assessment of sunflower seeds, oil and cake. Perspective of using sunflower oilcakes as a functional ingredient. Plants 10, 2487. doi: 10.3390/plants10112487
Podravac, D., Jager, N., and Lenart, L. (2018). “Microbiological quality analysis of organically grown oilseed products,” in Proceedings of the 11th Croatian Congress of Cereal Technologists, 21–30. Available online at: https://urn.nsk.hr/urn:nbn:hr:109:899699 (accessed February 14, 2023).
Porcellato, D., and Skeie, S. B. (2016). Bacterial dynamics and functional analysis of microbial metagenomes during ripening of Dutch-type cheese. Int. Dairy J. 61, 182–188. doi: 10.1016/j.idairyj.2016.05.005
Qi, Y., Huang, L., Zeng, Y., Li, W., Zhou, D., Xie, J., et al. (2021). Pediococcus pentosaceus: Screening and application as probiotics in food processing. Front. Microbiol. 12:762467. doi: 10.3389/fmicb.2021.762467
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Querol, A., Barrio, E., Huerta, T., and Ramon, D. (1992). Molecular monitoring of wine fermentations conducted by Active Dry Yeast strains. Appl. Environ. Microbiol. 58, 2948–2953. doi: 10.1128/aem.58.9.2948-2953.1992
Raak, N., Struck, S., Jaros, D., Hernando, I., Gülseren, I., Michalska-Ciechanowska, A., et al. (2022). Blending side streams. A potential solution to reach a resource efficient, circular, zero-waste food system. Fut. Foods 2022, 100207. doi: 10.1016/j.fufo.2022.100207
Raimondi, S., Anighoro, A., Quartieri, A., Amaretti, A., Tomás-Barberán, F. A., Rastelli, G., et al. (2015). Role of bifidobacteria in the hydrolysis of chlorogenic acid. MicrobiologyOpen 4, 41–52. doi: 10.1002/mbo3.219
Rao, M., Bast, A., and de Boer, A. (2021). Valorized food processing by-products in the EU: finding the balance between safety, nutrition, and sustainability. Sustainability 13, 4428. doi: 10.3390/su13084428
Risner, R., Tomasino, E., Hughes, P., and Meunier-Goddik, L. (2019). Volatile aroma composition of distillates produced from fermented sweet and acid whey. J. Dairy Sci. 102, 202–210. doi: 10.3168/jds.2018-14737
Rocha-Mendoza, D., Kosmerl, E., Krentz, A., Zhang, L., Badiger, S., Miyagusuku-Cruzado, G., et al. (2021). Acid whey trends and health benefits. J. Dairy Sci. 104, 1262–1275. doi: 10.3168/jds.2020-19038
Ryan, M. P., and Walsh, G. (2016). The biotechnological potential of whey. Rev. Environ. Sci. Biotechnol. 15, 479–498. doi: 10.1007/s11157-016-9402-1
Scharlack, N. F., Aracava, K. K., and Rodrigues, C. E. C. (2017). Effect of the type and level of hydration of alcoholic solvents on the simultaneous extraction of oil and chlorogenic acids from sunflower seed press cake. J. Sci. Food Agric. 97, 4612–4620. doi: 10.1002/jsfa.8331
Siedler, S., Balti, R., and Neves, A. R. (2019). Bioprotective mechanisms of lactic acid bacteria against fungal spoilage of food. Curr. Opin. Biotechnol. 56, 138–146. doi: 10.1016/j.copbio.2018.11.015
Skeie, S. B., Haland, M., Thorsen, I. M., Narvhus, J., and Porcellato, D. (2019). Bulk tank raw milk microbiota differs within and between farms: A moving goalpost challenging quality control. J. Dairy Sci. 102, 1959–1971. doi: 10.3168/jds.2017-14083
Tangyu, M., Muller, J., Bolten, C. J., and Wittmann, C. (2019). Fermentation of plant-based milk alternatives. Appl. Microbiol. Biotechnol. 103, 9263–9275 doi: 10.1007/s00253-019-10175-9
Terefe, N. S., and Augustin, M. A. (2020). Fermentation for tailoring the technological and health related functionality of food products. Crit. Rev. Food Sci. Nutr. 60, 2887–2913. doi: 10.1080/10408398.2019.1666250
USDA United States Department of Agriculture.. (2023). Oilseeds: World Markets and Trade. Foreign Agricultural Service. Available online at: https://www.fas.usda.gov/data/oilseeds-world-markets-and-trade (accessed March 20, 2023).
Vigentini, I., Praz, A., Domeneghetti, D., Zenato, S., Picozzi, C., Barmaz, A., et al. (2016). Characterization of malolactic bacteria isolated from Aosta Valley wines and evidence of psychrotrophy in some strains. J. Appl. Microbiol. 120, 934–945. doi: 10.1111/jam.13080
Von Gastrow, L., Amelot, R., Segond, D., Guézennec, S., Valence, F., and Sicard, D. (2021). Microbial community dispersal in sourdough. bioRxiv. doi: 10.1101/2021.10.18.464797
Wilmotte, A., Van der Auwera, G., and De Wachter, R. (1993). Structure of the 16S ribosomal RNA of the 325 thermophilic cyanobacterium Chlorogloeopsis HTF (Mastigocladus laminosus HTF) strain PCC7518 and 326 phylogenetic analysis. FEBS Lett. 317, 96–100. doi: 10.1016/0014-5793(93)81499-P
Winkler, H. (2011). Closed-loop production systems - A sustainable supply chain. CIRP J. Manufact. Sci. Technol. 4, 243–246. doi: 10.1016/j.cirpj.2011.05.001
Keywords: acid whey, food chain sustainability, Lactococcus lactis, Pediococcus pentosaceus, sunflower press cake, back-slopping technique, organic food
Citation: Mangieri N, Rosciano G, Porcellato D, Winther AR, De Noni I, Fracassetti D, Foschino R and Vigentini I (2023) Sustainability of food side streams: a case study of fermented blends made with sour whey and sunflower press cake powder using the back-slopping technique. Front. Sustain. Food Syst. 7:1166002. doi: 10.3389/fsufs.2023.1166002
Received: 14 February 2023; Accepted: 03 April 2023;
Published: 03 May 2023.
Edited by:
Guadalupe Virginia Nevárez-Moorillón, Autonomous University of Chihuahua, MexicoReviewed by:
Nitin Dhowlaghar, The University of Tennessee, Knoxville, United StatesAilton Cesar Lemes, Federal University of Rio de Janeiro, Brazil
Copyright © 2023 Mangieri, Rosciano, Porcellato, Winther, De Noni, Fracassetti, Foschino and Vigentini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Foschino, roberto.foschino@unimi.it
 Nicola Mangieri
Nicola Mangieri Gerardo Rosciano2
Gerardo Rosciano2  Davide Porcellato
Davide Porcellato Daniela Fracassetti
Daniela Fracassetti Roberto Foschino
Roberto Foschino Ileana Vigentini
Ileana Vigentini